| Issue 7 |
Cancer is one of the primary causes of death in the industrialised world. In the United Kingdom, one in four people will die of cancer, whilst one in three will be diagnosed with cancer at some point in their life. Whilst these figures make for grim reading, the arena of cancer research presents a wide range of challenges and opportunities for engineers. Engineers are set to play a key role in developing new approaches for the early detection of cancer, and for improving the treatment of cancer once it has been detected.
As a postdoctoral research fellow in biomedical engineering, I am working to advance the field of personalised cancer therapy (or more grandly - personalised medicine). Personalised cancer therapy is an emerging field of research which offers the potential for a dramatic improvement in the treatment and management of many common forms of cancer. The aim of this field is to develop techniques to enable specific, individualised treatment options for cancer patients. Thus, for a particular patient with a given tumour, personalised therapy involves determining the specific course of treatment (from the different options available) which will be most beneficial for the individual patient.
My current research is focused on two clinical studies which aim to advance the state-of-the-art in personalised cancer therapy. The first study, which is currently underway at the Churchill Hospital in Oxford, is concerned with the use of combined chemotherapy and radiation therapy (so-called chemoradiotherapy) for patients suffering from rectal cancer. This type of treatment is commonly given to patients prior to surgical removal of the tumour. The hope is that the treatment will cause the tumour to shrink or "downsize", which in turn will improve the likelihood of a more successful surgical resection and ultimately the long-term prognosis for the patient.
In reality the situation is more complex. Approximately 10% of rectal cancer patients treated with pre-operative chemoradiotherapy show a complete response, such that there is no visible tumour present in the resected specimen. Unfortunately, since the treatment response can only be evaluated after surgery, these "complete responders" undergo an unnecessary and highly invasive surgical procedure. At the other end of the spectrum, there are a significant number of patients (approximately 30%) who show little or no response to the treatment. For some of these patients, their tumours do not change much during the course of the treatment (so-called stable disease). However for others their tumours continue to grow in spite of the treatment, such that by the time of the surgery the cancer may be considerably more advanced than at the time of the initial diagnosis. For this subset of patients it would clearly be more beneficial for them to proceed directly to surgery, rather than undergoing a futile period of chemotherapy and radiation therapy. The problem for clinicians is that they currently have no way of knowing which patients will respond and which will not. The challenge for engineers is to develop accurate and robust techniques for predicting patient response from the available clinical data.
In order to address this problem, the patients in the rectal cancer study will undergo two different types of imaging scans, both before and after treatment with chemoradiotherapy. The first type of scan is known as dynamic contrast-enhanced magnetic resonance imaging, or DCE-MRI for short. With this type of imaging, the patient lies inside an MRI scanner and a small amount of a "contrast agent" is injected into the patient during the imaging procedure. The contrast agent is a specially designed paramagnetic molecule which is carried by the blood vessels throughout the body. The contrast agent arriving at the site of the tumour will leak into and back out of the "extravascular space" surrounding the tumour over a period of minutes. The resulting images, which are acquired at a temporal resolution of approximately ten seconds, can then be used to quantify the pattern of blood flow throughout the extent of the tumour. This can provide useful information regarding the aggressiveness of the given cancer, since more aggressive tumours typically develop a more advanced network of blood vessels.
The second type of imaging scan which the patients will undergo is known as positron emission tomography, or PET. With this approach, a radioactive probe or tracer is injected into the patient whilst they are lying inside a PET scanner. The counts due to decay of the radiolabelled tracer are then measured inside the scanner and this information is used to reconstruct a series of three-dimensional images of the concentration and location(s) of the tracer through time. The key to PET imaging is that the tracer molecule is designed in such a way that it becomes trapped inside metabolically-active cancer cells, yet is only taken up in relatively small amounts by normal healthy cells. Thus PET images can be used to quantify the metabolic activity of a given tumour, which in turn provides crucial information for predicting how the tumour will respond to different kinds of treatments.
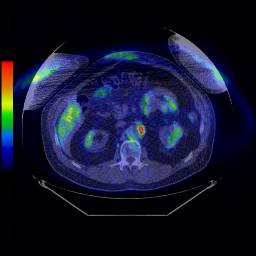
|
| Figure 1: PET/CT image for a patient with cancer. The "hot spot" in the image indicates the location of the tumour |
Given the DCE-MRI and PET scans for a particular patient, how do we then predict if the patient would benefit from pre-operative treatment with chemoradiotherapy, or whether it would be more beneficial for the patient to proceed directly to surgery? This is where techniques from the domain of information engineering can play a crucial role. Since modern imaging scans, such as those described above, produce 4-D data (3-D space + time), mathematical models must be used to extract compact and physiologically meaningful descriptions of the data.
In the case of DCE-MRI, we can use a type of model known as a "pharmacokinetic model" to quantify the rate constants which relate to the flow of the contrast agent into and out of the tumour. Similar models can be used with PET scans to quantify the rate constants describing the uptake of the tracer within the tumour. The parameters extracted from different types of imaging scans can then be combined or "fused" to generate a signature for a given tumour. A collection of such tumour signatures (for a large number of different patients) can then form the input to a machine learning algorithm, which can be used to identify the particular patterns that relate to the different classes of treatment response (e.g. complete response, stable disease, progressive disease).
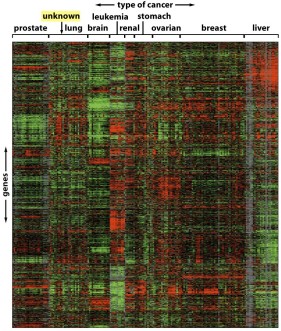
A similar approach to that described above is being employed in a separate study (also taking place at the Churchill Hospital) focused on a new drug for the treatment of breast cancer. The drug, which is known as Avastin, is designed to disrupt the network of blood vessels which feed the tumour (commonly referred to as the "tumour vasculature"). However, the response of breast cancer patients to Avastin is quite varied, with only one in five patients typically exhibiting a significant response. Since modern anti-cancer drugs such as Avastin are expensive and generally only benefit a subset of patients, there is a great deal of interest in identifying the markers that will predict those patients who will respond best in order to personalise these expensive therapies (see Figure 2).
|
|
Figure 2: Surveying the expression levels of thousands of genes in different
cancers using gene arrays. Higher-than-average levels of expression are indicated in red, whereas lower-than-average are in green |
To achieve this aim, we are using DCE-MRI scans to image the tumour vasculature before and after treatment with Avastin. The imaging data will then allow us to quantify the degree of vascular response to the drug for the different patients in the study. In addition, DNA microarrays or "gene arrays" are being used to examine the expression levels of thousands of genes within the tumours. Using this combined imaging and genomics approach, we hope to identify a gene expression signature that will enable clinicians to predict those patients whose tumours will be most responsive to Avastin. An additional benefit of this approach is that the particular set of genes which are predictive of response may also provide new insight into the biological mechanisms which differentiate responsive tumours from their non-responsive counterparts.
Although personalised approaches to cancer therapy such as those described above offer the potential for a substantial change in the way in which cancer patients are treated, a more dramatic change may come from the advent of early detection technologies1. A significant research effort is currently underway to identify biomarkers - trace amounts of proteins produced by cancers - which can be detected reliably and robustly within small blood samples. By identifying multiple biomarkers for different tumour types, it should be possible to develop simple diagnostic tests which can be used to detect cancer at an early stage. Imaging, and in particular molecular imaging approaches such as PET scans, can then be used to identify the location of the nascent tumour within the body. Once the tumour has been localised, early-stage intervention procedures can be used to treat the cancer whilst it is still in its infancy.
The motivation for the early detection approach is simple: the chances of patient survival are significantly greater if cancer is diagnosed whilst still confined to the organ of origin and before it has had a chance to spread to other parts of the body. Conversely, survival rates tend to decrease dramatically as tumours enlarge and subsequently metastasise.
As with personalised cancer therapies, early detection strategies will require solutions to a number of key engineering challenges if they are to come to fruition. How can we build accurate and reliable models for predicting the presence of an early-stage cancer given (noisy) measurements of multiple biomarkers from blood samples? How can we accurately localise small early-stage tumours using medical imaging scanners with limited spatial resolution? And how can we improve the characterisation of a given tumour by fusing the multiple sources of information which are increasingly available (i.e. multi-modal imaging scans, protein biomarker measurements, and gene expression profiles)? These challenges and more will continue to keep engineers working on cancer busy for many years to come.
| << Previous article | Contents | Next article >> |
| SOUE News Home |
Copyright © 2008 Society of Oxford University Engineers |
SOUE Home |